Abstract
Background: Angioimmunoblastic T-cell lymphoma and other nodal lymphomas of T follicular helper (TFH) cell origin (TFH lymphoma) have a poor prognosis and are associated with immune dysregulation. However, comprehensive immune profiles of TFH lymphoma have not been fully investigated. The RHOA G17V mutation (G17V) is a disease-specific mutation found in 60-70% of TFH lymphoma and considered to gain a novel function through binding to VAV1 protein. Nevertheless, the prevalence of homozygous (Homo) and heterozygous (Hetero) G17V mutations and the difference in their oncogenicity due to its zygosity have not been elucidated. Here, we performed single-cell transcriptomic analysis to characterize tumor and immune cells of TFH lymphoma.
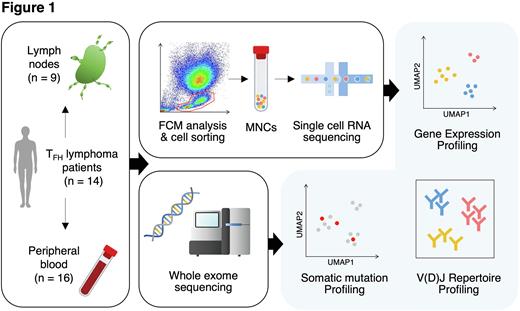
Methods: We performed single-cell RNA sequencing (scRNA-seq) and T-cell/B-cell receptor sequencing (scTCR/BCR-seq) for 9 lymph nodes (LNs) and 16 peripheral blood (PB) from 14 TFH lymphoma patients (9 at initial diagnosis [ID] and 5 at relapsed/refractory diagnosis [RR]), as well as 7 homeostatic LNs (HLNs) from patients with solid cancers (Fig. 1). Whole-exome sequencing (WES) was also performed for genetic mutation analysis using bulk DNA extracted from tumor tissues and PB of the same cohort as scRNA-seq. scRNA-seq and scTCR/BCR-seq libraries were constructed using the Chromium system (10x Genomics). Publicly available scRNA-seq data from 5 healthy donors (HDs) were also integrated as control. scRNA-seq data analysis was performed by Seurat, Scater, and Batchelor for quality control, integration, and clustering; SingleR for annotation; Immunarch for TCR/BCR repertoire analysis; gene set variation analysis (GSVA) for pathway analysis, and CellPhoneDB for cell-cell interaction analysis. Genomon2 pipeline was used for calling somatic mutations in WES data. Subsequently, VarTrix was applied to estimate mutations detected by WES in scRNA-seq data.
Results: In total, 68,738 cells from LNs and 98,891 cells from PB were analyzed. Unsupervised clustering and annotation by canonical markers identified 4 major cell types: CD4/CD8+ T, natural killer (NK), B, and myeloid cells. Tumor cells were identified by the expanded TCR clonality and the TFH-like gene expression signatures. Next, using scRNA-seq data, we reanalyzed a total of 393 somatic mutations discovered by WES and identified 15 mutations in more than 5 cells of scRNA-seq data, of which only RHOA G17V was identified in multiple samples. G17V-mutated cells were detected in 4 LN and 6 PB samples, mostly restricted to tumor cells. G17V-Homo and Hetero cells were identified based on frequency of mutant reads. Differentially expression gene (DEG) analysis and GSVA in G17V-mutated tumor cells of LNs revealed that the regulatory signature was significantly activated in Homo, whereas the proliferation signature was up-regulated in Hetero. Additionally, DEG analysis identified three tumor-specific gene candidates and the expression of cell surface protein PLS3 was validated by flow cytometry and immunohistochemistry. Subsequently, we performed sub-clustering and GSVA of immune cells and found that dysfunctional CD8 T cells were significantly increased in LNs and PB of patients compared to controls and exhibited oligoclonal expansion by TCR repertoire analysis. Notably, TREG, complement component 1q positive macrophages (C1Q+ Mφ), and LAMP3+ dendritic cells (DCs) were significantly increased in LNs from patients with RR diseases than HLNs. DEG analysis and GSVA revealed high expression of PD-L1 and regulatory signatures in LAMP3+ DCs. Moreover, genes related to immune suppression were significantly upregulated in TREG of patients compared to those of controls. Finally, cell-cell interaction analysis uncovered that tumor cells and TREG of RR patients strongly interacted with C1Q+ Mφ and LAMP3+ DCs via CXCR3-CXCL9 and CCR4-CCL17 interactions, respectively, indicating that these myeloid cells not only induced TREG into tumor tissues but also actively interacted with tumor cells.
Conclusion: Tumor cells of TFH lymphoma acquired the immunosuppressive signature in the clonal evolution process from the G17V Hetero to Homo. Furthermore, comprehensive immune profiling of TFH lymphoma revealed expansion of cells involved in the immune escape. These results unveiled the enrichment of immune evasive phenotypes in TFH lymphoma and suggested their contribution to treatment resistance in RR patients.
Disclosures
Suzuki:Daiichi Sankyo Co., Ltd: Research Funding. Chiba:Kyowa Kirin Co., Ltd.: Research Funding; Chugai Pharmaceutical Co., Ltd.: Research Funding; Thyas Co., Ltd.: Research Funding; Bayer Yakuhin Ltd.: Research Funding; Eisai Co., Ltd.: Research Funding; Astellas Pharma Inc.: Research Funding. Sakata-Yanagimoto:Otsuka Pharmaceutical Co., Ltd.: Research Funding; Eisai Co., Ltd.: Research Funding; Bristol-Myers Squibb Company: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal